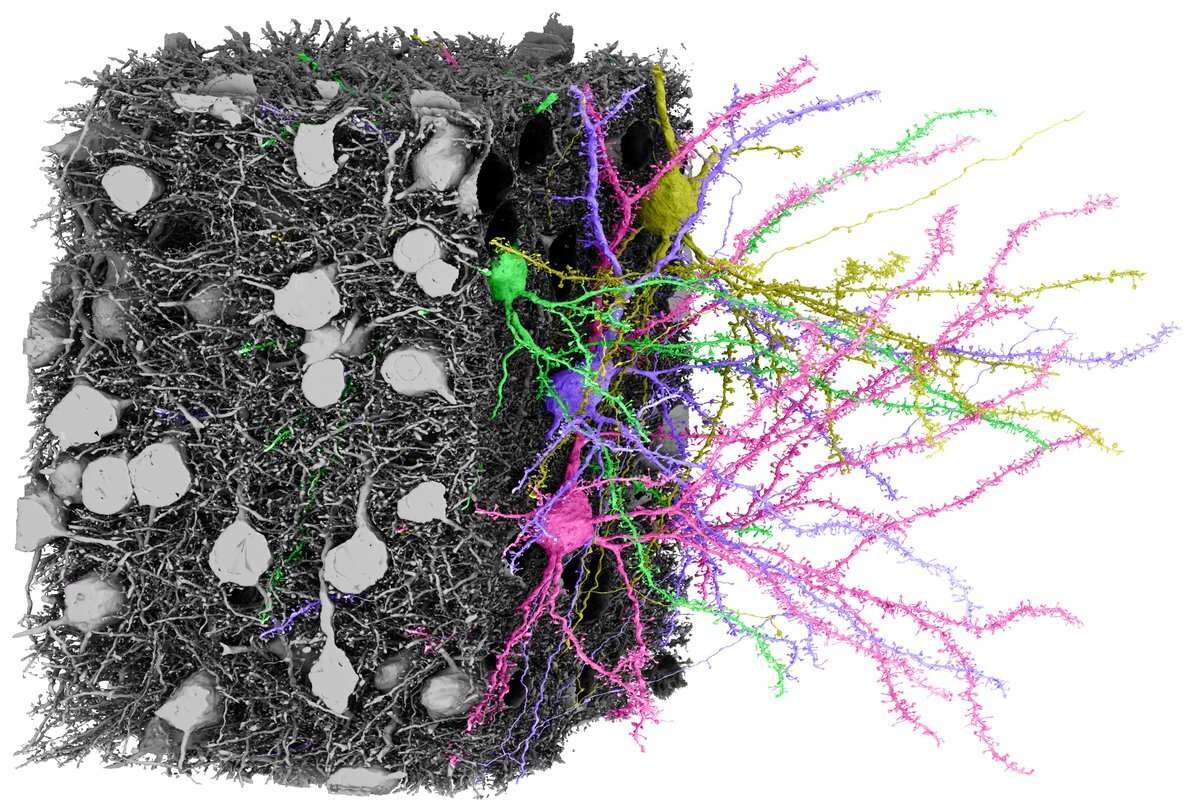
Explore EM data from layer 2/3 of the mouse visual cortex
This dataset was the first acquired and analyzed by the initial phase of the project. It is a 250 x 140 x 90 µm volume from layer 2/3 of a P36 male mouse visual cortex imaged at 3.58 x 3.58 x 40 nm resolution with a dense segmentation, proofreading of all dendrites and axons of the 364 excitatory neurons in the volume and a dense synapse detection.
As an ongoing experiment, we have made a publicly available version of the dataset available for anyone to proofread and dynamically analyze, under the CAVE datastack name “pinky_sandbox”. All data or proofreading contributed to this dataset is fully public.
Access
We are releasing datasets in versions. Each individual version resembles a state of the segmentation during proofreading. Release notes contain detailed documentation on the nature of the tables and state of the dataset for each release.
Release v185.4
Technical descriptions of state of the dataset, proofreading, and known errors in this version.
Interactive Visualization
You can visualize and interact with the dataset via neuroglancer (an online data visualization tool).
Basic neuroglancer instructions are here.
Dataset available at this link. (supports Chrome or Firefox)
NOTE: If you are interested in large scale downloading of the data, please contact info@microns-explorer.org. There are real costs to the project that are incurred doing this and there are efficient and inefficient ways to do this.
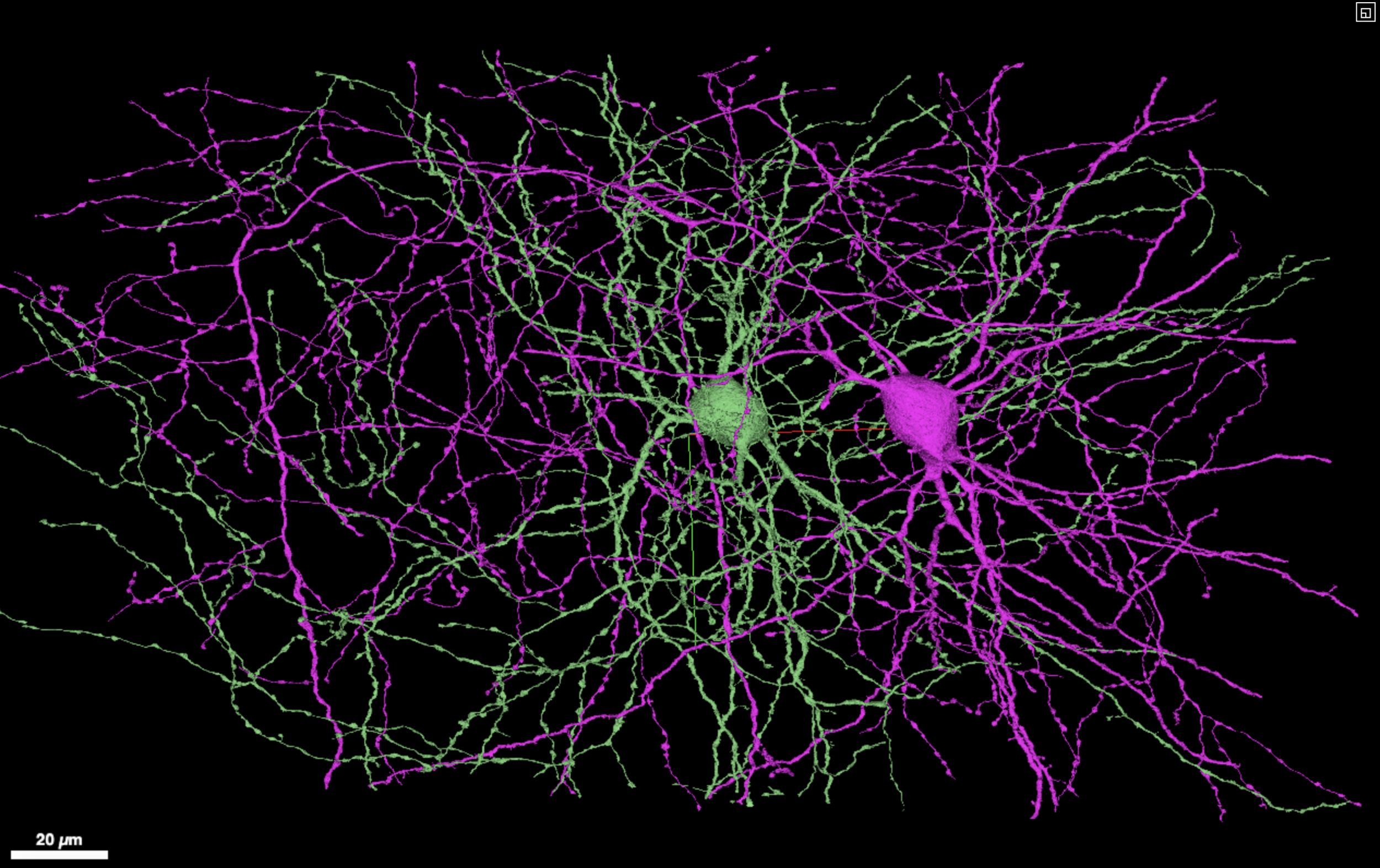
A screenshot of an in browser visualization of two inhibitory neurons with extensive axonal reconstructions.
Synapse Table
This is a table of 3.2 million rows of putative synapses between objects within the volume. It was generated by an automated synapse detection approach (Turner et al 2019). Many of the objects in the dataset are not proofread, and so this cannot be trivially interpreted as a connectivity graph. [link]. Note: contains uint64 values and will not display properly in Excel. Note this was updated on 1/6/2022 to make the IDs properly match the synapse cleft segmentation IDs.
Soma Valence Table
This csv contains the spatial locations of most of the cells with somas in the volume, as well as indication of whether that cell is an excitatory cell, an inhibitory cell, or a glial cell of some type. It furthermore identifies the neurons for which proofreading has been complete, and the soma subgraph for which the analysis in Dorkenwald et al 2019 was based. [link]. Note: contains uint64 values and will not display properly in Excel. This was updated on 1/6/2022 to add the nucleus_id in as an available column.
Fixed Meshes of Cells with Soma in the dataset
This set of h5 files contain numpy arrays of mesh vertices, faces, and extra edges that connect disconnected portions of the mesh. (link) These files can be read using MeshParty [documentation).
Proofread Soma Subgraph Synapses
This contains a csv file of synapses which have been further proofread to remove false positive synapses between the excitatory cells with somas in the volume. It also contains the spine head volume measurements. This is the smallest file necessary to recapitulate the analysis in Dorkenwald et al., 2019 (link) Note: contains uint64 values and will not display properly in Excel.
Functional Data
The functional data is available in a processed form.
The visual stimulus shown to the animal is available as zipped movies here.
The functional traces from cells after extracting calcium events as described in the EASE publication can be found here as a pandas pickle file here.
The structural two photon stack taken from the mouse in vivo before the EM is available here.
The co-registration points between the structural stack and the EM volume is available here as a csv, and an accompanying python notebook demonstrating how to access and utilize them.
There is also a readme document with more details about the functional data.
The raw two photon movies (90GB) are available upon request (info@microns-explorer.org).
Examples of using these data products can be found in the Microns Binder repository.
Skeletons
We have contributed a subset of the cells to the Brain Imaging Library in swc skeleton format. You read about and download that data from this location on brain-map.org
Cell Classifications
A human labelled coarse cell classification for all objects with soma in the volume is available at this location.
Use the soma valence table (here) to link these to nuclei.
Ground Truth Data
We have released the ground truth data that was used to train the various machine learning models for this dataset. You can download the data here (3.4 GB).
For a complete list of data products available for this dataset and a unique DOI see the below Zenodo link.
Mitochondria Ground Truth
Manual mitochondria annotations are available at the following google bucket:
precomputed://gs://neuroglancer/pinky10/ground_truth_registration/dst/mit_new
See the segmentation in neuroglancer here.
Papers to Cite the Dataset
Structural EM data and segmentation
Binary and analog variation of synapses between cortical pyramidal neurons.
Dorkenwald, S., Turner, N.L., Macrina, T., Lee, K., Lu, R., Wu, J., Bodor, A.L., Bleckert, A.A., Brittain, D., Kemnitz, N., et al. (2019). bioRxiv 2019.12.29.890319; doi: https://doi.org/10.1101/2019.12.29.890319
Structure and function of axo-axonic inhibition
Schneider-Mizell, C. Bodor, A.L., Collman, F. Brittain,D. Bleckert, AA, Dorkenwald, S., Turner N.L. Macrina, T. Lee, K. Lu, R. Wu, J. et al. eLife 2021;10:e73783 DOI: 10.7554/eLife.73783
Reconstruction of neocortex: Organelles, compartments, cells, circuits, and activity
Nicholas L. Turner*, Thomas Macrina*, J. Alexander Bae*, Runzhe Yang*, Alyssa M. Wilson*, Casey Schneider-Mizell*, Kisuk Lee1*, Ran Lu*, Jingpeng Wu*, Agnes L. Bodor*, Adam A. Bleckert*, Derrick Brittain*, Emmanouil Froudarakis*, Sven Dorkenwald*, Forrest Collman*, Nico Kemnitz* et al.
Cell, Volume 185, Issue 6, 2022, Pages 1082-1100.e24, https://doi.org/10.1016/j.cell.2022.01.023.
Functional imaging data
Zhou, P. et al. EASE: EM-Assisted Source Extraction from calcium imaging data. bioRxiv 2020.03.25.007468; doi: https://doi.org/10.1101/2020.03.25.007468
Contributions
Functional Imaging: Emmanouil Froudarakis, Jacob Reimer, Andreas Tolias
Co-registration: Nuno Maçarico da Costa, Adam Bleckert, Marc Takeno
Tissue Preparation: Joann Buchanan, Marc Takeno, Nuno Maçarico da Costa
Sectioning: Agnes Bodor, Adam Bleckert
TEM Design and Maintenance: Derrick Brittain, Clay Reid
TEM Operation: Derrick Brittain, Wenjing Yin, Adam Bleckert, Marc Takeno, Daniel Bumbarger, Nuno Da Costa
EM Stitching and Rough Alignment: Gayathri Mahalingam, Russel Torres, Yang Li, Thomas Macrina, Dodam Ih
EM Fine Alignment: Thomas Macrina, Will Wong
Neuron segmentation: Kisuk Lee, Jingpeng Wu
Agglomeration: Ran Lu
Synapse Detection: Nicholas Turner, Jingpeng Wu
Neuroglancer Frontend: Nico Kemnitz, Manuel Castro
Proofreading Backend: Sven Dorkenwald, Nico Kemnitz, Chris S. Jordan
Cloud Data Interface: William Silversmith, Ignacio Tartavull
Connectome Versioning System: Forrest Collman, Sven Dorkenwald, Casey Schneider Mizell
Proofreading: Agnes Bodor, Nuno Maçarico da Costa, Szi-Chieh Yu, Alyssa Wilson, Sven Dorkenwald, Casey Schneider-Mizell, Forrest Collman
Project Management: Lynne Becker, Shelby Suckow
Scientific Management: Clay Reid, Nuno Da Costa, Andreas S. Tolias, Jacob Reimer, H. Sebastian Seung
IARPA MICrONS Program Management: David A. Markowitz, Jacob Vogelstein